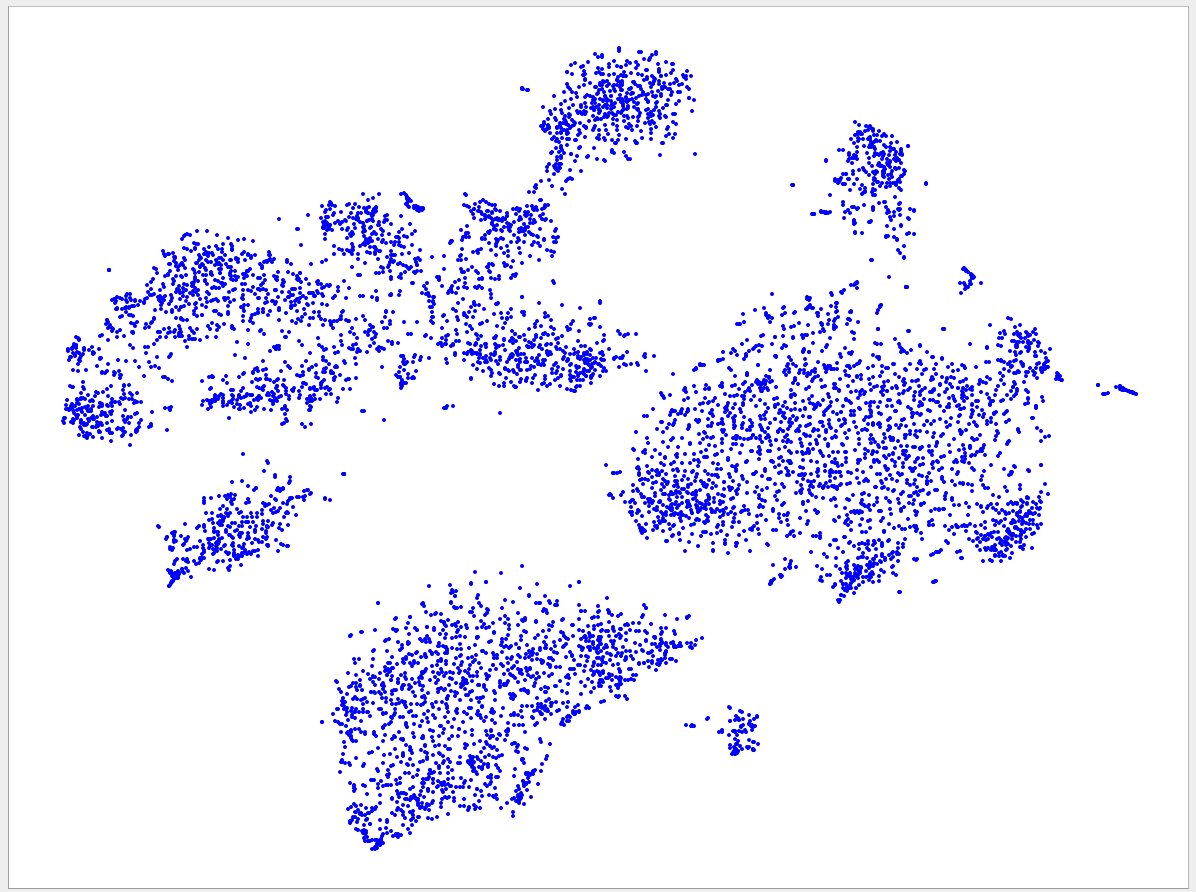
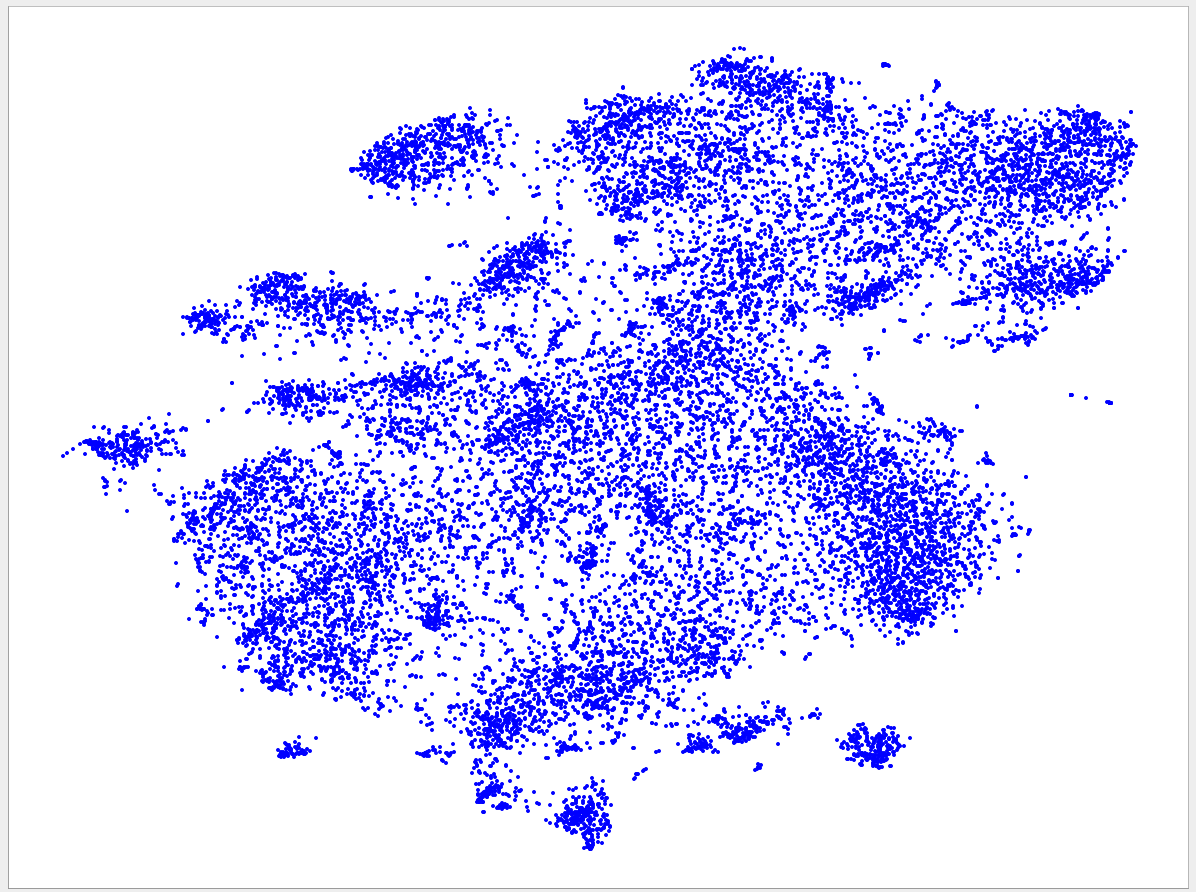
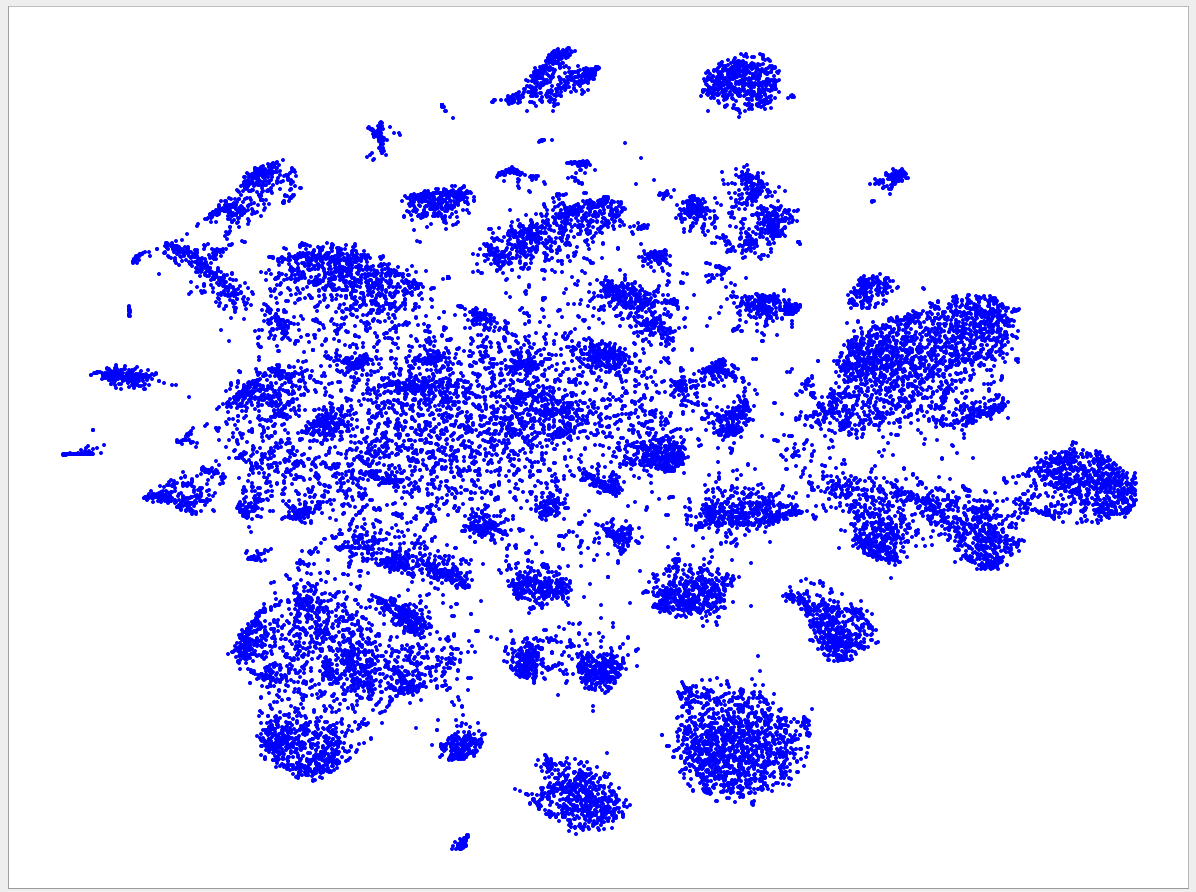
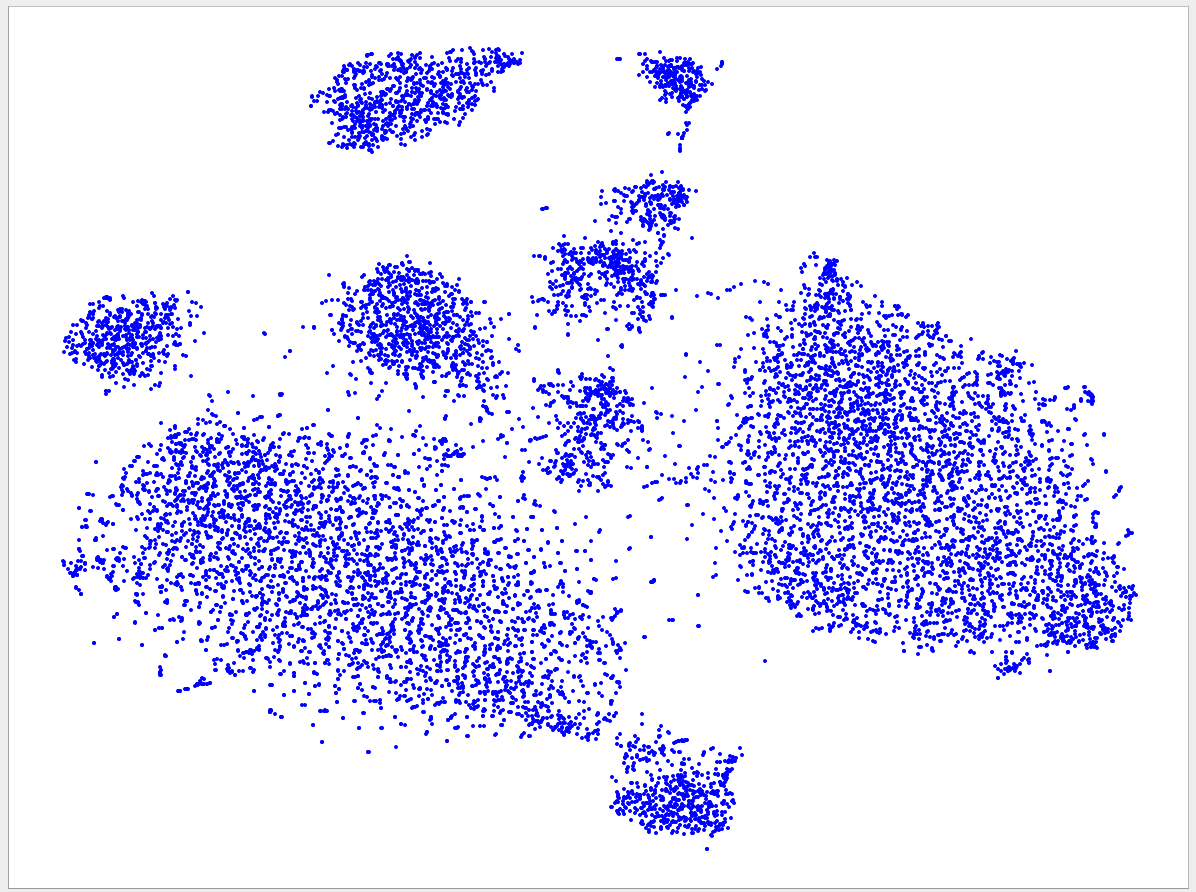
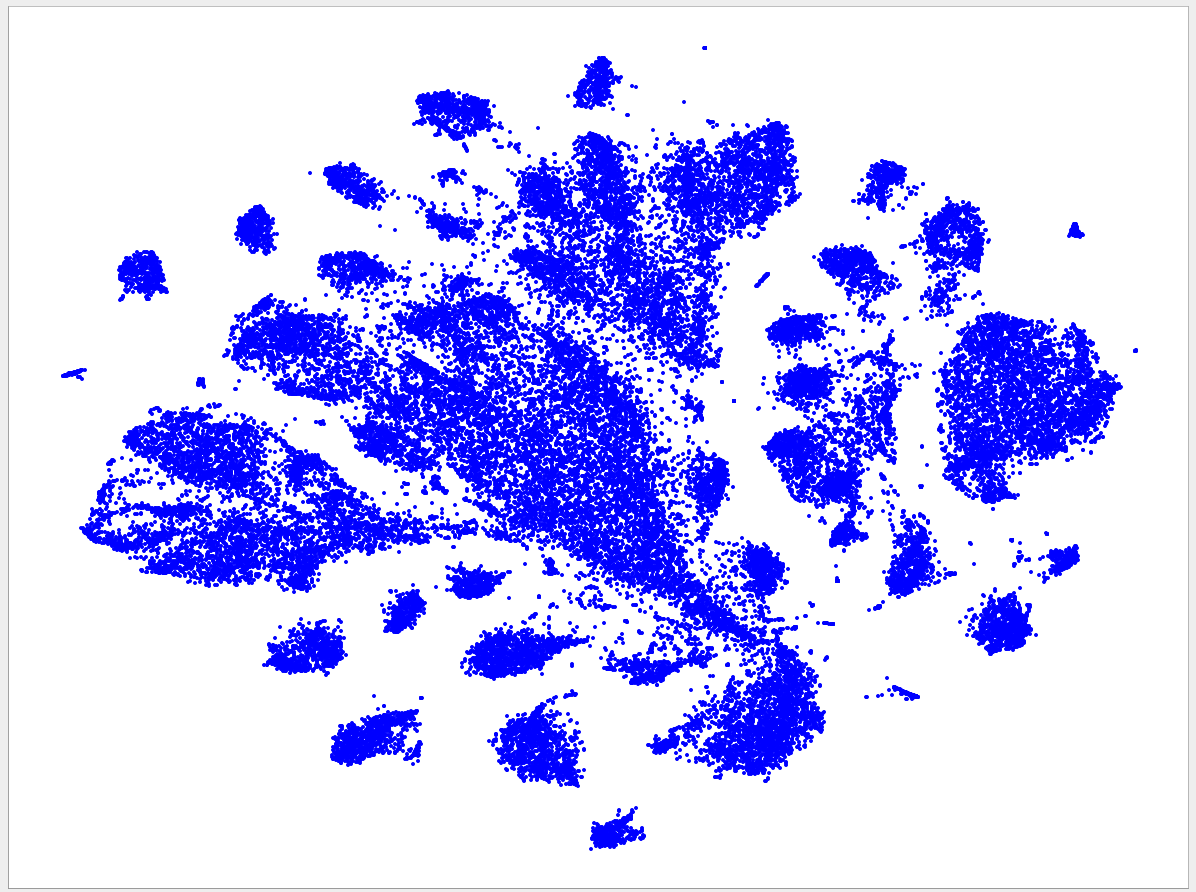
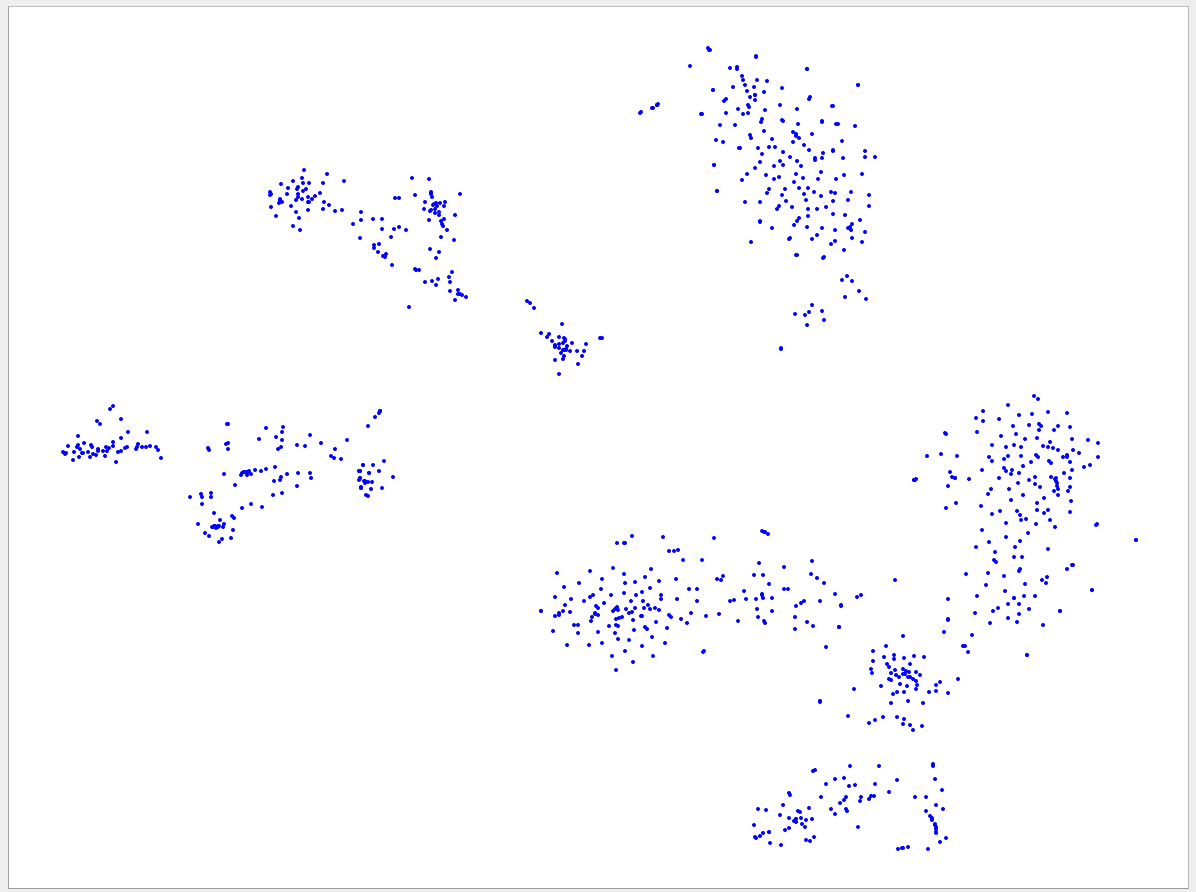
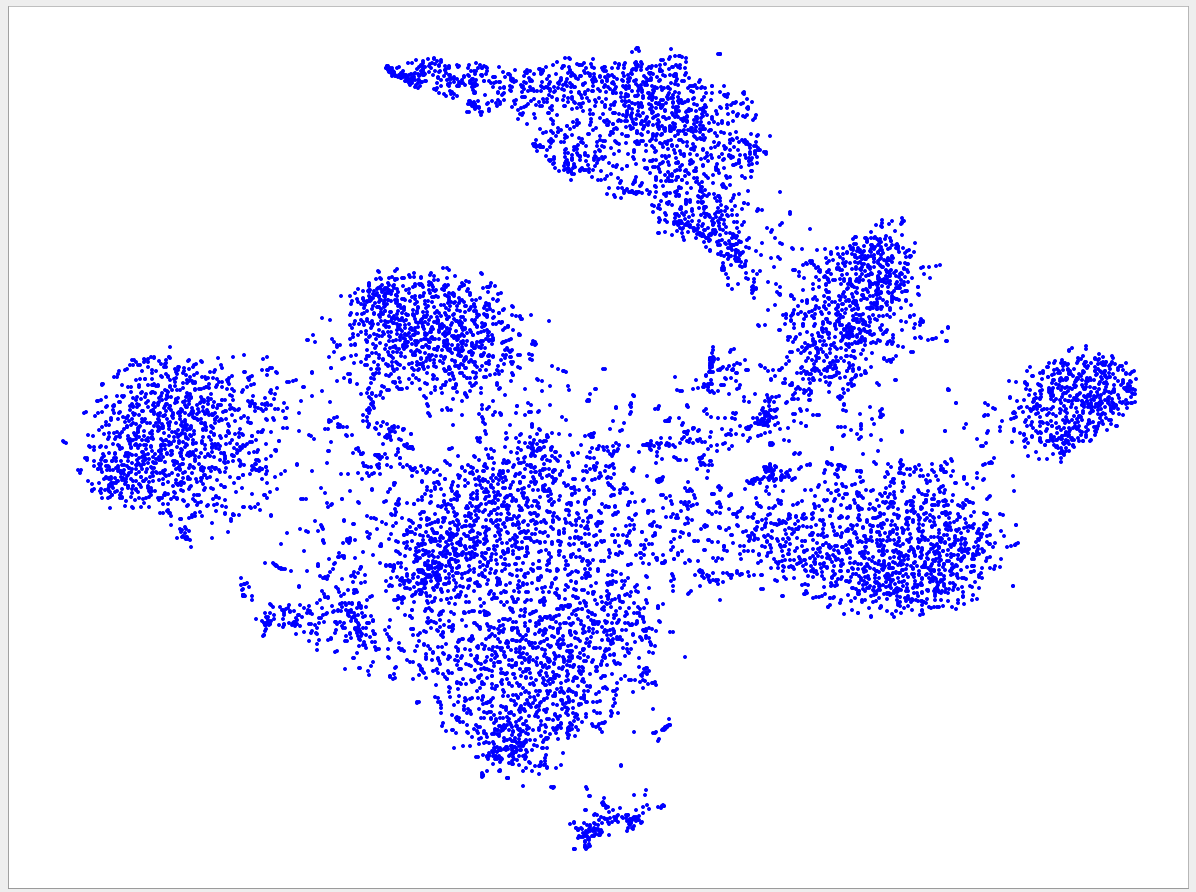
Welcome to the VizBin gallery page.
You can find 2D embeddings, screenshots, and runtimes of a collection of published metagenomic datasets here. More datasets may be included in the future
Should there be any particular metagenomes that you would like to add/see added, please let me know by opening an issue. Your feedback is greatly appreciated.
Stay tuned for updates!
If not stated otherwise, 4 threads and 3GB of memory (-Xmx3g) were used.
Visualizations of more recent assemblies usually have a lower minimum length threshold than for older assemblies The lower the threshold the more genomic content is expected to be recovered. Typically, the more recent assemblies result in better separated clusters and thus allow the inclusion of sequences of a lower minimum length. One reason for the better separation might be that more recent assemblies use longer reads and/or improved assemblers and thus produce better contigs. "Better" means here that the genomic signatures of the contigs are closer to the genomic signature of the originating population-level genome. Hence, less "spurious" signatures are introduced. This, in turn, is expected to lead to better separated clusters.
Acknowledgments
Many people have collaborated on making this application become a reality: Cedric C. Laczny, Tomasz Sternal, Valentin Plugaru, Piotr Gawron, Arash Atashpendar, Hera Houry Margossian, Sergio Coronado, Laurens van der Maaten, Nikos Vlassis, and Paul Wilmes.
The present work was supported by an ATTRACT programme grant (A09/03) and a European Union Joint Programming in Neurodegenerative Diseases grant (INTER/JPND/12/01) to Paul Wilmes and an Aide à la Formation Recherche grant (AFR PHD/4964712) to Cedric C. Laczny all funded by the Luxembourg National Research Fund (FNR).